17 March 2023
Application of Multi-Omics technologies for tree improvement
Jie Huang, Teagasc Walsh Scholar, and Dr Dheeraj Rathore, Teagasc Tree Improvement Researcher, discuss the potentials of omics technologies in accelerating the tree improvement research.
Tree breeding programmes have been developed for commerciallly and ecologically important tree species throughout the world for the purpose of developing improved genotypes that are suitable for afforestation, reforestation and conservation under climate change. They have been successfully delivering improved plant material with traits of interest such as higher yield & quality timber, better stem form, and higher tolerance to a/biotic stresses.
Traditionally, tree breeding techniques involve identification, selection and clonal propagation of ‘plus’ trees – trees seleted for their good phenotypic traits – which have been intensively developed and are commonly used in forest tree improvement programmes. However, these methods are time consuming and slow to breed trees due to their long generation cycle. In recent years, Multi-OMICS technologies such as genomics have emerged as promising tool to accelerate the tree breeding process by shortening the generation intervals. Moreover, studying the genetic adaptation to changing climate, and exploring the genomic basis of these changes has also become possible.
What are OMICS technologies?
OMICS technologies are integrative methods, which employ large volume of data representing the structure and function of an entire makeup of a given biological system to provide comprehensive understanding on growth and development of organisms and their response to (a)biotic changes. OMICS usually refer to genomics, transcriptomics, proteomics, metabolomics, phenomics and ionomics, with the first four being the most common ones. Together with bioinformatics and machine-learning approaches, OMICS have advanced our understanding of the growth and development of trees.
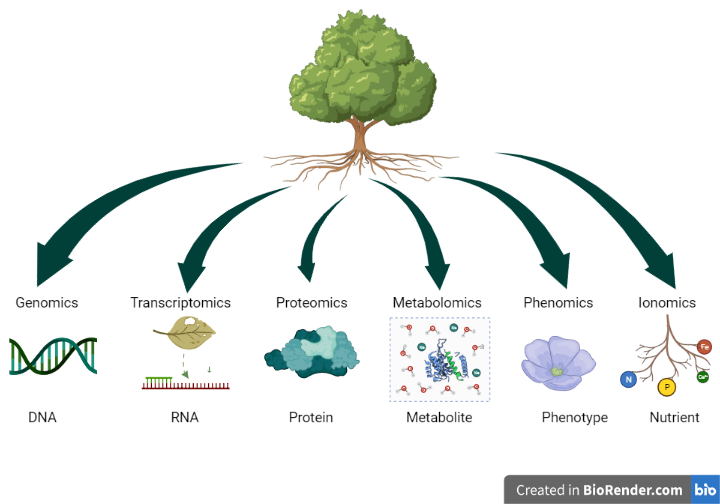
Figure 1: Building blocks of OMICS technologies
Did you know?
Genomics is the study of the entire genetic material of an organism including structure and function of genes and genomes. Genomics have given insights into genetic variation, sequence polymorphism, chromosomal organization, as well as enabling the identification of traits of interest through reconstruction of genetic maps.
Transcriptomics is the study of all RNA transcripts that are produced by a genome of an organism in a cell or tissue. Transcriptomics allows us to understand the gene expression patterns of an organism at its different developmental stages and/or in response to any biotic and abiotic stress over a certain period.
Proteomics is the study of total produced proteins in an organism and their interactions. Protein is the product from gene expression and proteomics would allow us to understand the localization, structures & functions, interaction of the produced protein over a certain period.
Metabolomics is the comprehensive measurement of all the metabolites and low molecular weight molecules in an organism. Metabolomics allows us to detect alterations in the biological pathways by measuring its response to anomalous processes such as those associated with disease.
Phenomics is the study of characterization of phenotypes on an organism-wide scale. Phenomics allows us to understand the relationship between phenotypic (or physical) traits and genetic variation.
Ionomics is the study of the complete cellular inorganic components in an organism. It allows us to investigate how organism respond to the physiological and environmental changes through distribution, composition and accumulation of ionome.
Tree improvement and OMICS
OMICS technologies have been successfully applied in tree improvement programmes for commercially and ecologically important tree species. Genomics help breeders to select and breed genetically improved trees through gene function identification, genomic breeding and gene editing. Genetic variation is vital for the adaptability, survival and sustainability of a forest, especially for monoculture restorative forests that are fragile from biotic and abiotic stress (e.g. invasive pathogen). Using the information from genomics and phenomics, Genome-Wide Association Study (GWAS) and Quantitative Trait Locus (QTL) analysis could demonstrate association to increase the efficacy of genomic selection of advantageous traits.
Association studies on Sitka spruce have successfully demonstrated locuses responsible for budburst and height. Moreover, a gene has been identified in silver birch to be responsible for early senescence and increased sugar levels associated with plant-microbes and/or plant-pest interactions by multi-omics approaches including genomics, transcriptomics, metabolomics and phenomics. Trees have large chemically complex mechanisms that they employ as a major defense system against abiotic stress and pathogens. For instance, European common ash (Fraxinus excelsior) has been severely threatened by the invasive fungal pathogen Hymenoscyphus fraxineus, known as Ash Dieback. A study of metabolomics on a variety of ash individuals with different level of susceptibility to ash dieback demonstrated that a total of 64 candidate metabolites were found to be associated with susceptiblity of ash to ash dieback. Furthermore, a study using transcriptomics has identified molecular markers for tolerance of ash to ash dieback.
Current research
Teagasc has established in-situ (e.g., ‘natural’ field sites) and ex-situ (e.g., nurseries) conservation gene banks and seed orchards for commercially important broadleaved tree species consisting of the full collection of ‘plus’ trees for alder, silver & downy birch and sycamore.
Broadgen is a Teagasc-funded project on broadleaf geneticsstarted in January 2022 and focuses on assessing the genetic diversity and resilience of commercially important broadleaved tree species in Ireland. This research project, funded by Teagasc Walsh Scholarship Programme, is a collaborative project with Prof Trevor Hodkinson in Trinity College Dublin, Dr Colin Kelleher in National Botanic Gardens, Dr Susanne Barth, Dr Stephen Byrne and Dr Dheeraj Rathore in Teagasc. The proposed research will provide baseline knowledge to support afforestation/reforestation programmes with higher tolerance to pathogens by:
- Assessing genetic diversity and population structure of alder, downy birch, silver birch, and sycamore from gene banks and seed orchards in Ireland,
- Exploring the susceptibility of ash and alder to fraxineus and Phytophthora alni, respectively, by inoculation experiments, and phenotyping the disease severity in ash gene-banks under field conditions,
- Associating the molecular markers related to Ash Dieback-tolerance and phenotypic traits such as height, diameter, time of bud flush, time of leaf shedding and severity of disease in ash genotypes from different provenances.
For further information on the BroadGen project, please visit https://www.teagasc.ie/crops/forestry/research/broadgen-project/

